Arvores de Regressão - XGBoost
Bibliotecas
Avaliando, selecionando dados
Treino e Teste com todas as variáveis
## Vamos criar os conjuntos de treino teste e desenvolver a arvore
## com todas as variáveis.
library(caret)
set.seed(2025)
indice <- createDataPartition(dados$medv, times=1, p=0.75, list=FALSE)
conj_treino <- dados[indice,]
conj_teste <- dados[-indice,]
str(conj_treino)'data.frame': 381 obs. of 14 variables:
$ crim : num 0.0273 0.0299 0.0883 0.1446 0.17 ...
$ zn : num 0 0 12.5 12.5 12.5 12.5 12.5 12.5 0 0 ...
$ indus : num 7.07 2.18 7.87 7.87 7.87 7.87 7.87 7.87 8.14 8.14 ...
$ chas : int 0 0 0 0 0 0 0 0 0 0 ...
$ nox : num 0.469 0.458 0.524 0.524 0.524 0.524 0.524 0.524 0.538 0.538 ...
$ rm : num 6.42 6.43 6.01 6.17 6 ...
$ age : num 78.9 58.7 66.6 96.1 85.9 94.3 82.9 39 61.8 84.5 ...
$ dis : num 4.97 6.06 5.56 5.95 6.59 ...
$ rad : int 2 3 5 5 5 5 5 5 4 4 ...
$ tax : num 242 222 311 311 311 311 311 311 307 307 ...
$ ptratio: num 17.8 18.7 15.2 15.2 15.2 15.2 15.2 15.2 21 21 ...
$ black : num 397 394 396 397 387 ...
$ lstat : num 9.14 5.21 12.43 19.15 17.1 ...
$ medv : num 21.6 28.7 22.9 27.1 18.9 15 18.9 21.7 20.4 18.2 ...str(conj_teste)'data.frame': 125 obs. of 14 variables:
$ crim : num 0.00632 0.02729 0.03237 0.06905 0.21124 ...
$ zn : num 18 0 0 0 12.5 0 0 0 0 75 ...
$ indus : num 2.31 7.07 2.18 2.18 7.87 8.14 8.14 8.14 8.14 2.95 ...
$ chas : int 0 0 0 0 0 0 0 0 0 0 ...
$ nox : num 0.538 0.469 0.458 0.458 0.524 0.538 0.538 0.538 0.538 0.428 ...
$ rm : num 6.58 7.18 7 7.15 5.63 ...
$ age : num 65.2 61.1 45.8 54.2 100 36.6 94.1 85.7 88.8 21.8 ...
$ dis : num 4.09 4.97 6.06 6.06 6.08 ...
$ rad : int 1 2 3 3 5 4 4 4 4 3 ...
$ tax : num 296 242 222 222 311 307 307 307 307 252 ...
$ ptratio: num 15.3 17.8 18.7 18.7 15.2 21 21 21 21 18.3 ...
$ black : num 397 393 395 397 387 ...
$ lstat : num 4.98 4.03 2.94 5.33 29.93 ...
$ medv : num 24 34.7 33.4 36.2 16.5 20.2 15.6 13.9 14.8 30.8 ...Preparando os dados
x_treino <- model.matrix(medv ~ . , data = conj_treino)[, -1]
y_treino <- conj_treino$medv
x_teste <- model.matrix(medv ~ . , data = conj_teste)[, -1]
y_teste = conj_teste$medv
dtrain <- xgb.DMatrix(data = x_treino, label = y_treino)
dtest <- xgb.DMatrix(data = x_teste, label = y_teste)Treinamento com validação cruzada e grid de parâmetros
set.seed(2025)
# Grid de hiperparâmetros
grid <- expand.grid(
eta = c(0.01, 0.1),
max_depth = c(3, 6),
nrounds = c(100, 200, 300)
)
# Avaliar cada combinação com CV
resultados_cv <- list()
for (i in 1:nrow(grid)) {
params <- list(
objective = "reg:squarederror",
eta = grid$eta[i],
max_depth = grid$max_depth[i],
verbosity = 0
)
cv <- xgb.cv(
params = params,
data = dtrain,
nrounds = grid$nrounds[i],
nfold = 5,
metrics = "rmse",
early_stopping_rounds = 10,
verbose = 0
)
resultados_cv[[i]] <- list(
rmse = min(cv$evaluation_log$test_rmse_mean),
best_nrounds = cv$best_iteration,
params = grid[i, ]
)
}
# Melhor modelo
rmses <- sapply(resultados_cv, function(x) x$rmse)
melhor_indice <- which.min(rmses)
melhor_param <- resultados_cv[[melhor_indice]]$params
melhor_nrounds <- resultados_cv[[melhor_indice]]$best_nrounds
melhor_rmse <- resultados_cv[[melhor_indice]]$rmse
melhor_param eta max_depth nrounds
6 0.1 3 200melhor_nrounds[1] 122melhor_rmse[1] 3.198383Modelo Final
Importancia das variáveis
# Importância das variáveis
importance_matrix <- xgb.importance(model = final_model)
# Gráfico
xgb.plot.importance(importance_matrix)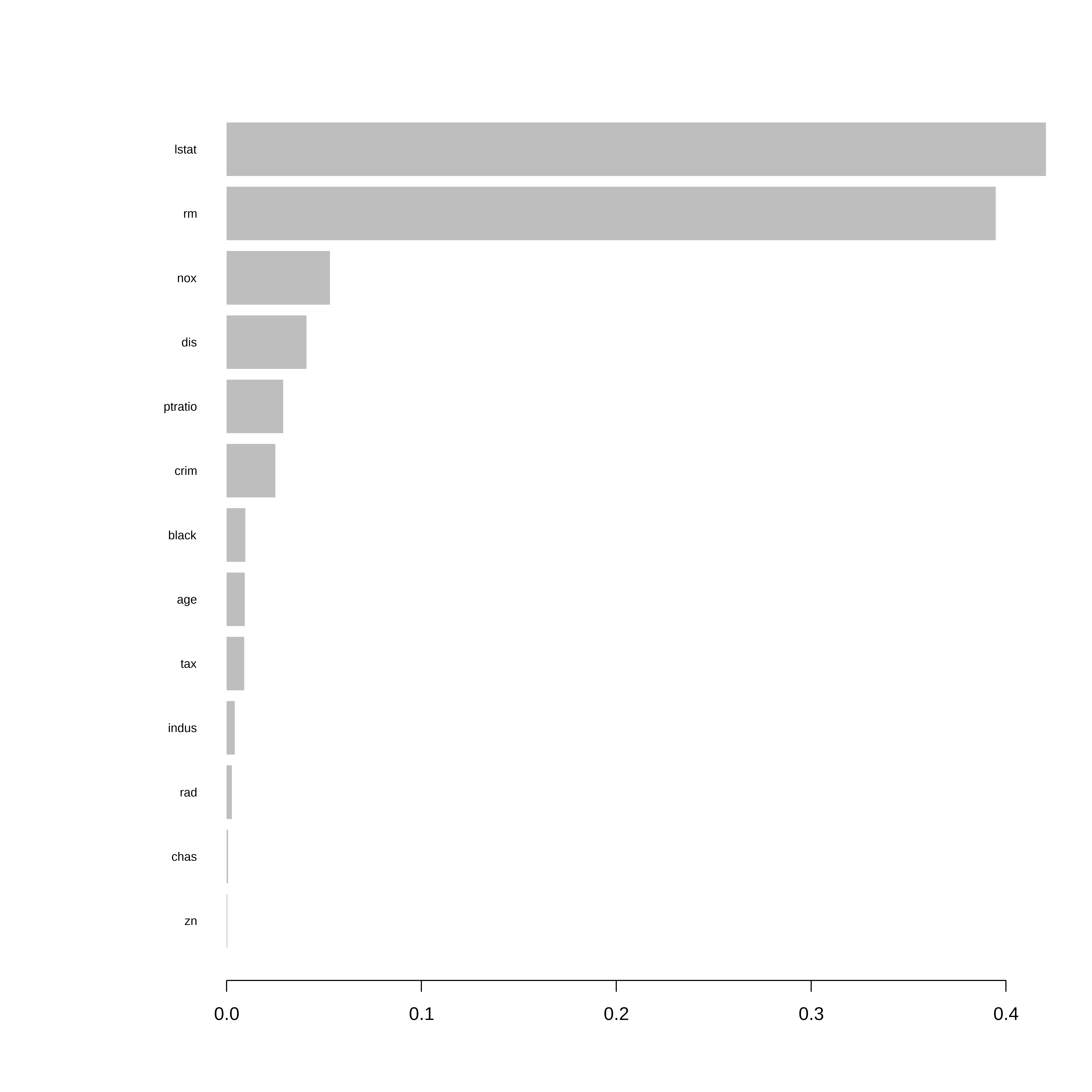
Previsões
conj_teste$prev <- predict(final_model, dtest)
ggplot(conj_teste, aes(x = prev, y = medv)) +
geom_point() +
geom_abline()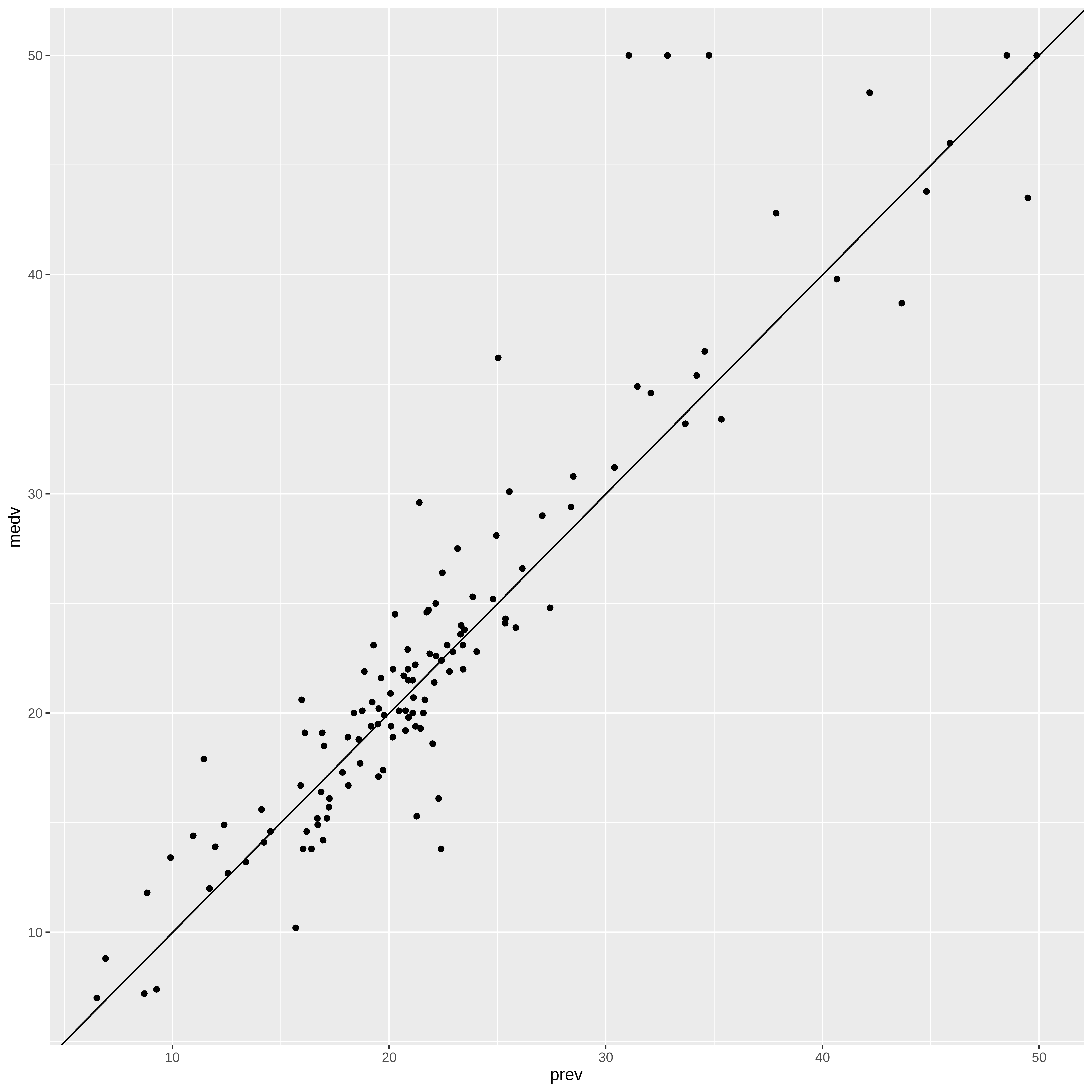
Calculando o RMSE
RMSE no conjunto de teste: 3.34077caret::postResample(conj_teste$prev, conj_teste$medv) RMSE Rsquared MAE
3.3407695 0.8947614 2.1166538 Comparação com outro modelo (Regressão Linear)
ctrl <- trainControl(method = "cv", number = 5)
model_lm <- train(
medv ~ ., data = conj_treino,
method = "lm",
trControl = ctrl
)
pred_lm <- predict(model_lm, newdata = conj_teste)
postResample(pred_lm, conj_teste$medv) RMSE Rsquared MAE
5.5535662 0.7120235 3.8617314